Overview
- Task: Segment cerebral vasculature on contrast-enhanced CT by background subtraction.
- Imaging modality: any, with baseline (non-contrasted) and contrasted images available.
Prerequisites
- SlicerElastix and SlicerVMTK extensions are installed.
- Slicer-4.10 or later.
Recommended workflow
- Align baseline (non-contrast) CT and contrast-enhanced CT using
General registration (Elastix)module to minimize artifacts due to patient motion. - Subtract the baseline volume from contrast-enhanced volume using
Subtract scalar volumesmodule. - Use
Vesselness filteringmodule to remove non-vessel shaped structures (bone plates that have not been fully suppressed by subtraction). - In
Segment editormodule, use vesselness filtered image as master volume and extract vessels usingThresholdeffect. - Remove remaining artifacts using
Islandseffect’sKeep largest islandmethod. - Optionally compute vessel centerline using
Centerline extractionmodule.
Tips:
- If there is only rigid patient motion between baseline and contrast images then
generic rigid (all)registration preset may be used, which attempts to align the images by applying rigid transform. It may be more robust and faster than using a warping transform. - Find a module by hitting Ctrl + F and start typing its name
- Adjust window/level of the image to improve visibility of vessels in slice viewers: click Window/Level mouse mode in the toolbar and Ctrl + Left-click-and-drag with the mouse (starting from a vessel point) to set optimal window/level automatically.
Example
- Load
CTABrainBaselineandCTABrainContrastvolumes (get the images using the links below). Significant patient motion is visible. To minimize artifacts, we need to align the images before subtraction.
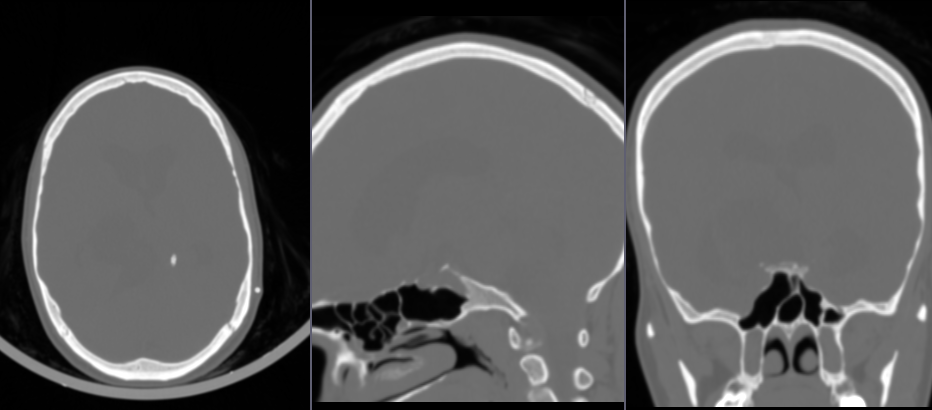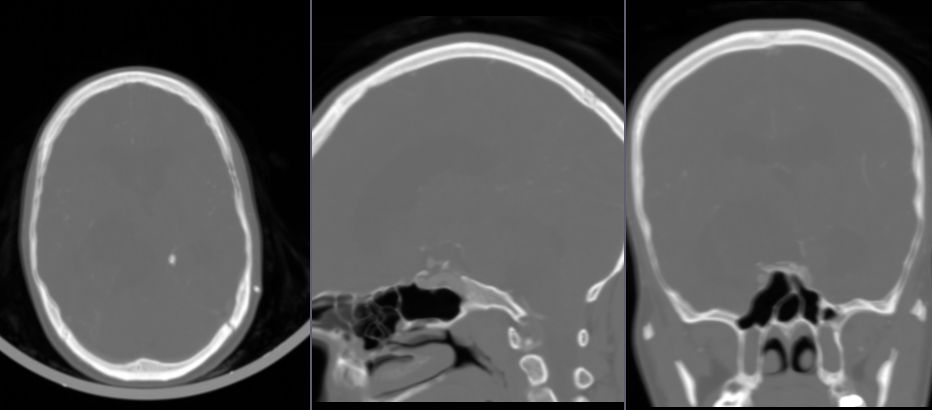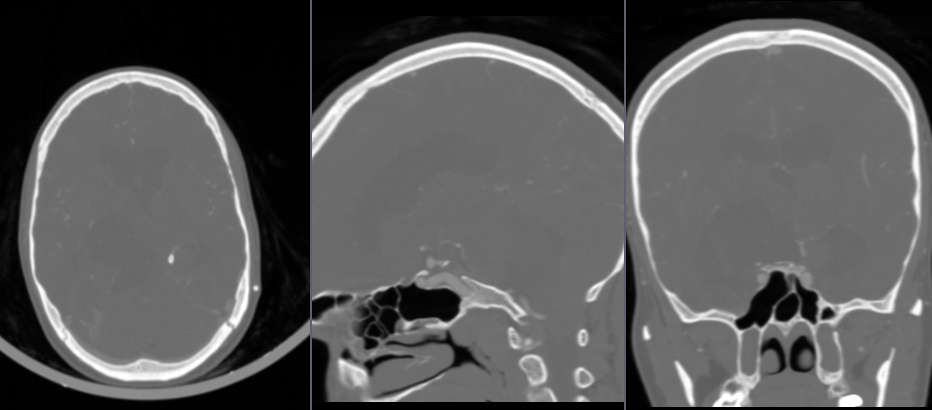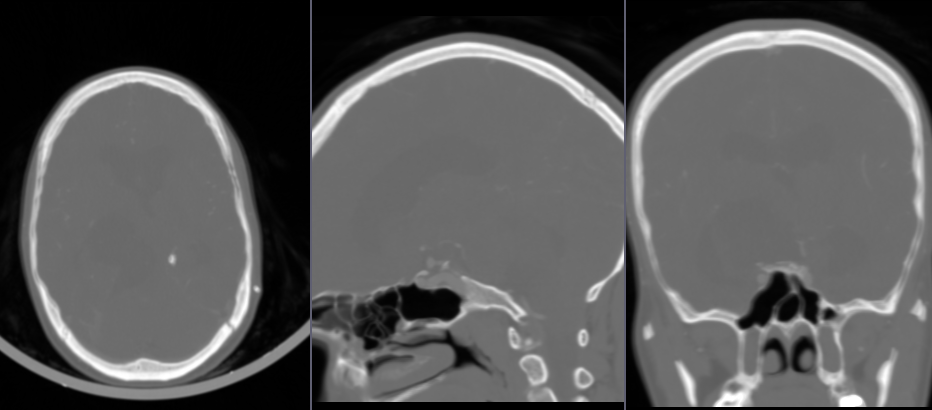
- Go to
General registration (Elastix)module - For
Fixed volumechoose baseline (no-contrast) volume - For
Moving volumechoose contrast volume as - For
Presetchoosegeneric (all) - For
Output volume, chooseCreate new Volume as...and typeCTABrainContastAligned - Click
Applyand wait until the registration completes (it may take several minutes)
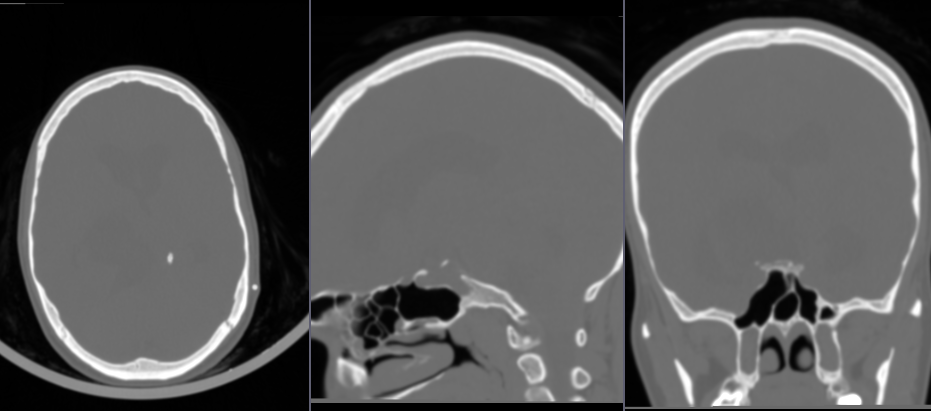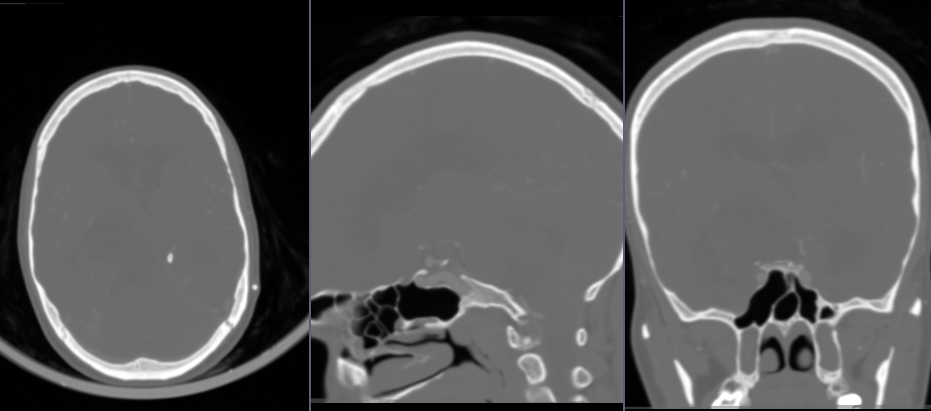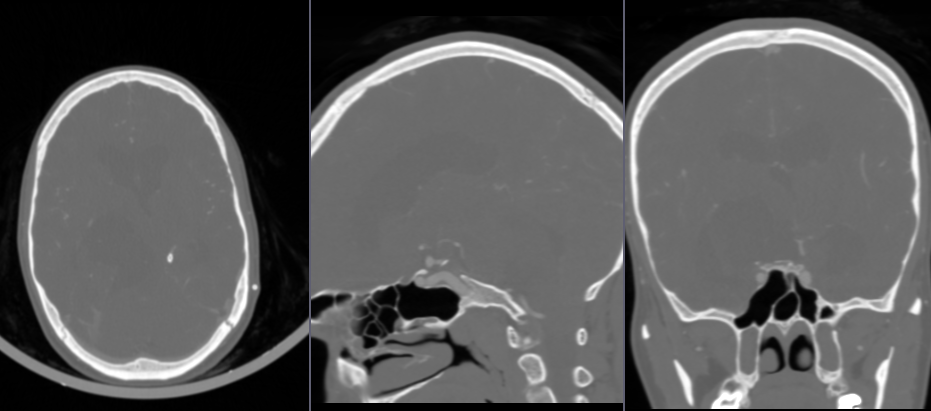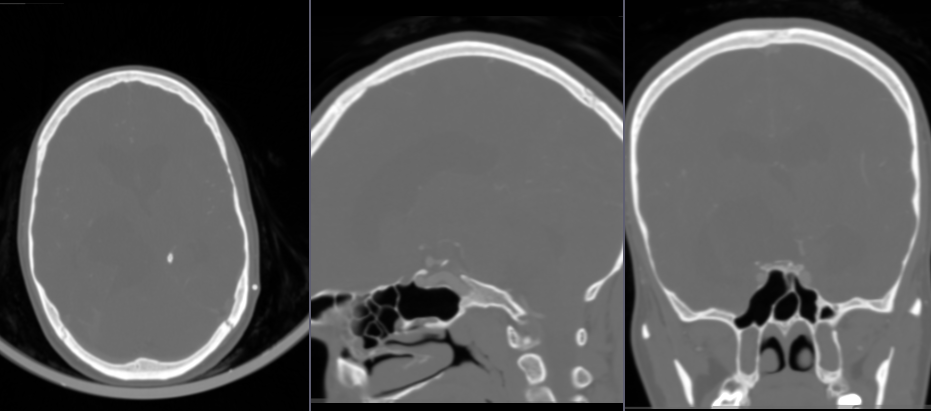
- Go to
Subtract scalar volumesmodule to subtract baseline image from contrast image - For
Input volume 1chooseCTABrainContrastAligned - For
Input volume 2chooseCTABrainBaseline - For
Output volume, chooseCreate new Volume as...and typeCTABrainSubtracted - Click
Apply
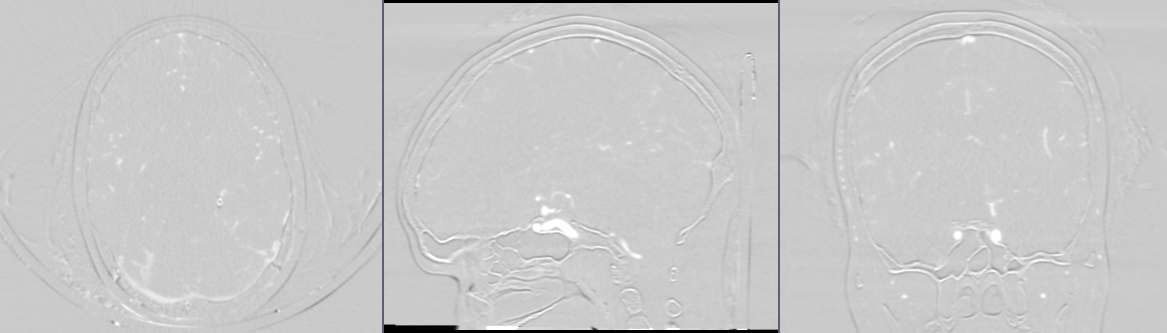
- Go to
Vesselness filteringmodule - For
Input volumechooseCTABrainSubtracted - For
Seed pointchooseCreate new MarkupsFiducialand click in a vessel point in the image. Tip: Choose a point that is not trivial to segment, for example one that is near bone surfaces that are not fully suppressed by the subtraction. - Click
Preview, wait for the filter preview to complete.
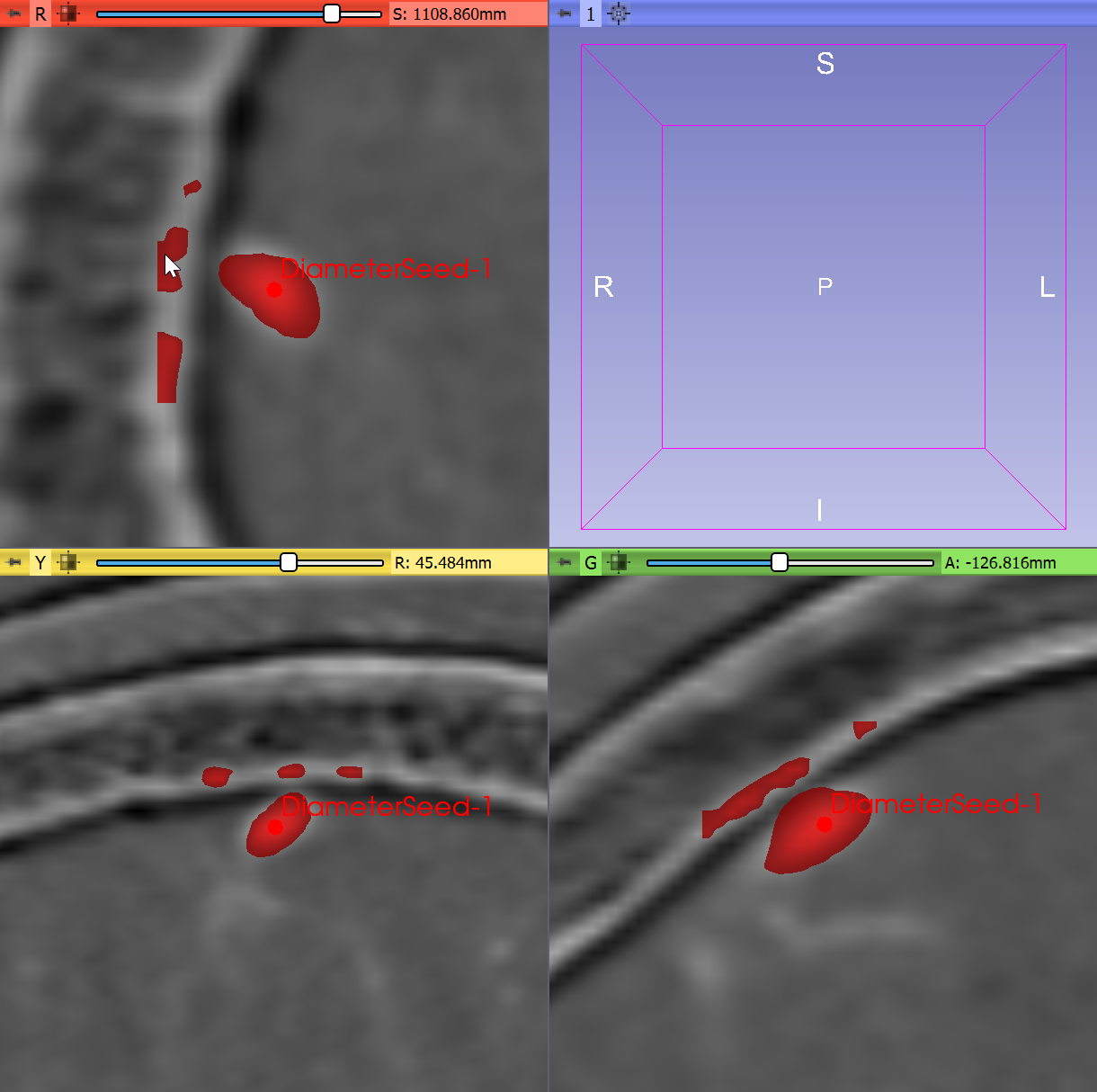
- Zoom out the slice views and inspect the results: some parts of the bone is detected as vessel. Increase suppression of plate-like structures by opening
Advancedsection and increasingSuppress platesvalue to 35% (other values that are used in this example:Minimum vessel diameter= 1 voxels,Maximum vessel diameter= 5 voxels,Vessel contrast= 52,Suppress blobs= 10%).
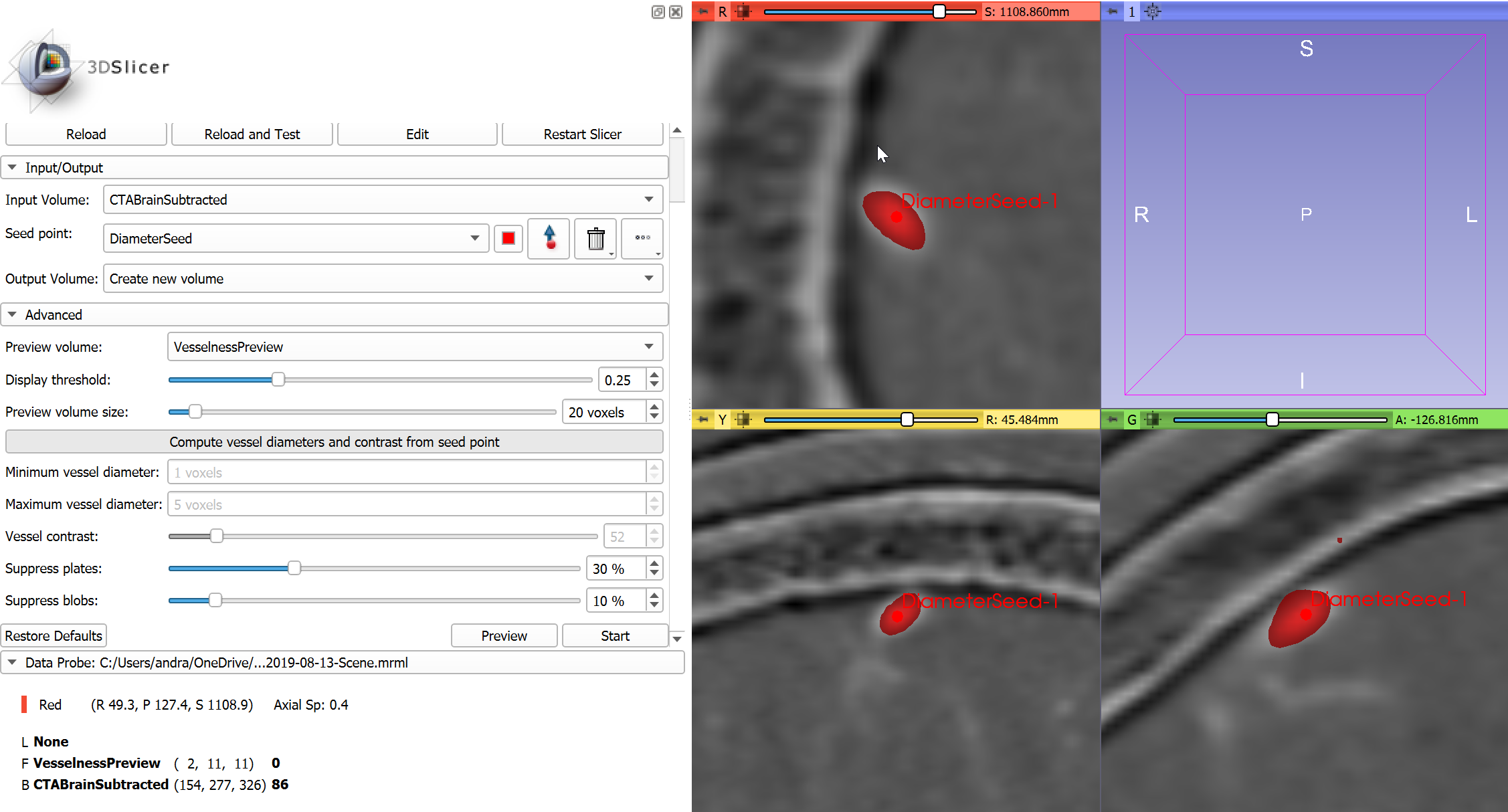
- Optional: Check filtering results at a few different locations by deleting current point, then placing a new point in the slice viewer (click the trashcan icon next to
Seed point, click the arrow button next to it, and click in the image). ClickPreviewbutton to see filtering results. -
Click
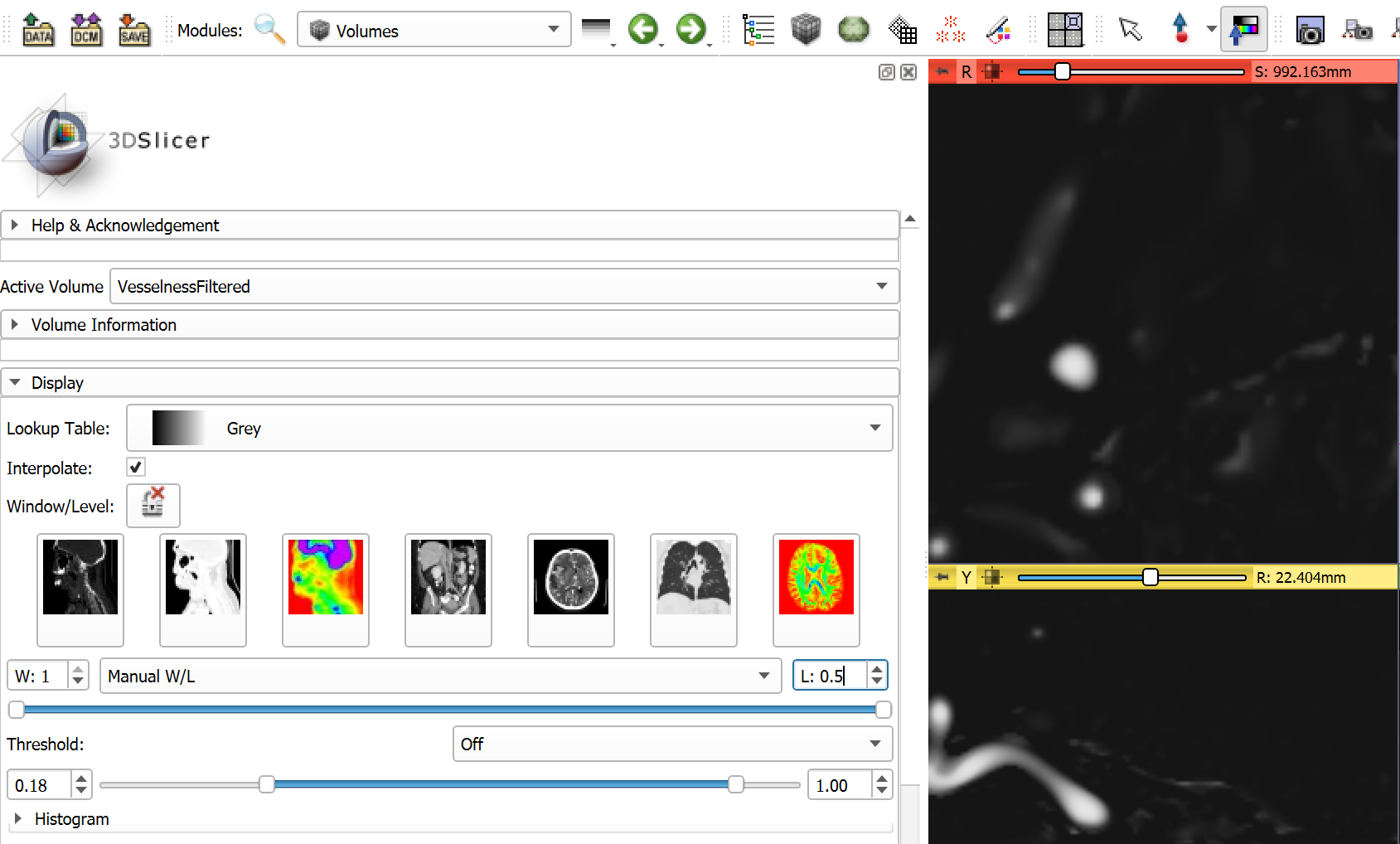
Startto perform vesselness filtering on the entire image. The operation may take a few minutes. - Optional: Adjust visualization of vesselness image in Volumes module. Set window (
W) value to 1.0, set level (L) value to 0.5. SetThresholdtoOff.
- Go to
Segment editormodule. - Click
Addbutton to create a new segment. - Double-click on the green rectangle in the
Colorcolumn, start typingArteryand hitEnterto accept the proposed name and color. - Select Threshold effect, set lower threshold value to 0.7 - to show the relevant vessels but still keep amount of artifacts under control. Click
Apply.
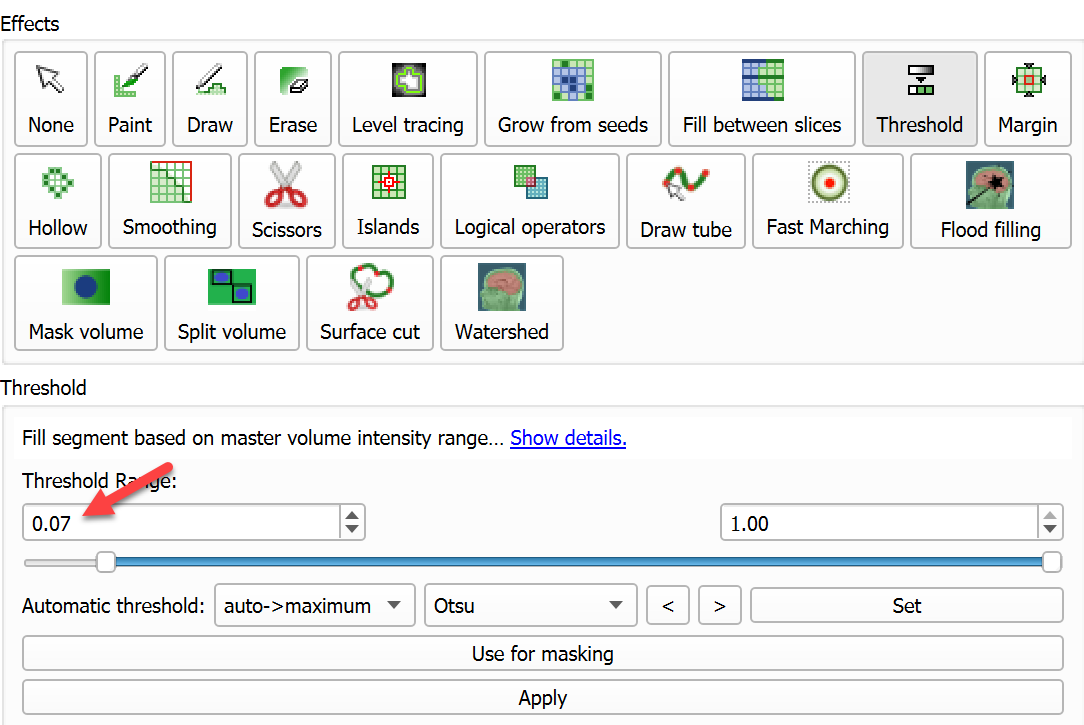
- Visualize the results in 3D: Uncheck
Surface smoothingin dropdown menu ofShow 3Dbutton to make 3D display update faster and prevent shrinking of vessels. Then clickShow 3D buttonto see the vessels in the 3D view.
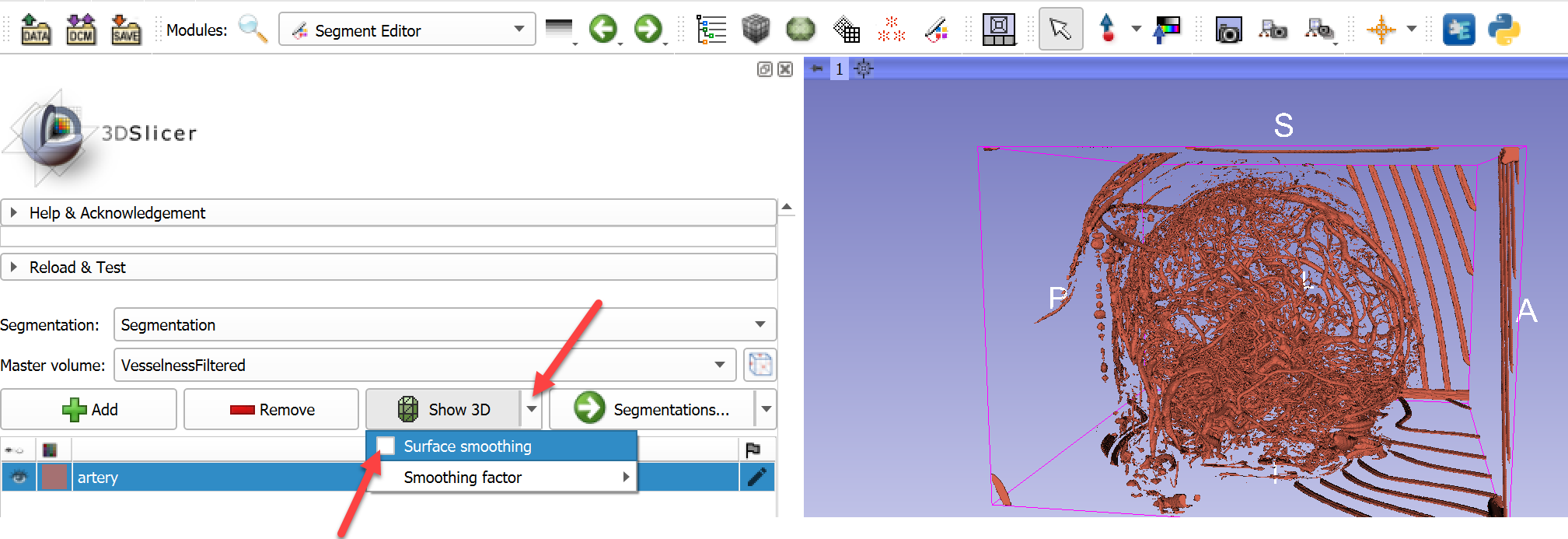
- To remove disconnected, non-vascular structures, click
Islandseffect, use the defaultKeep largest islandoption, and clickApply.
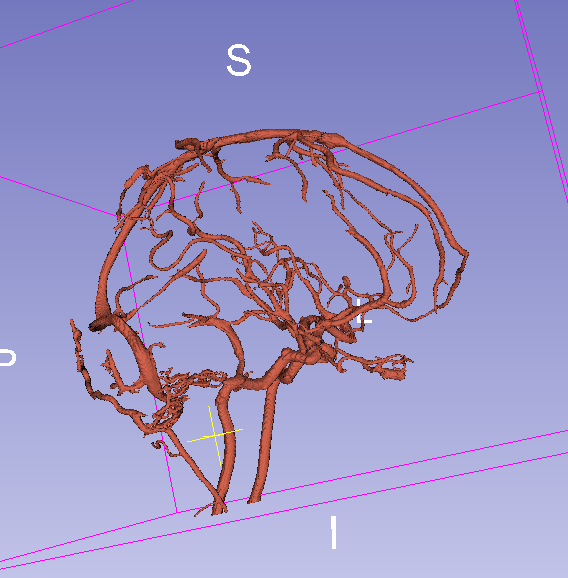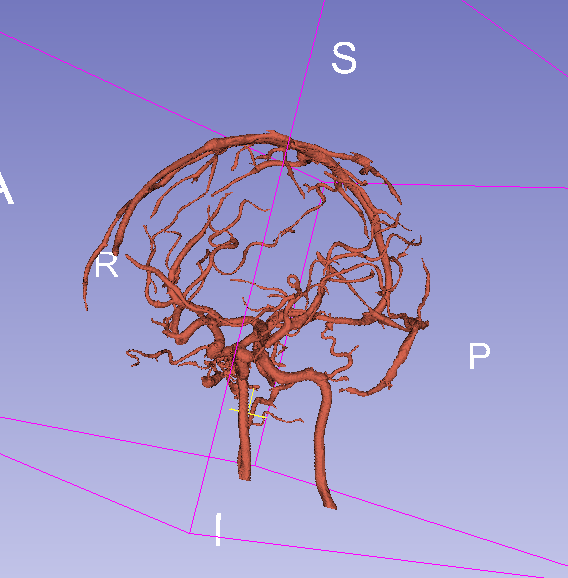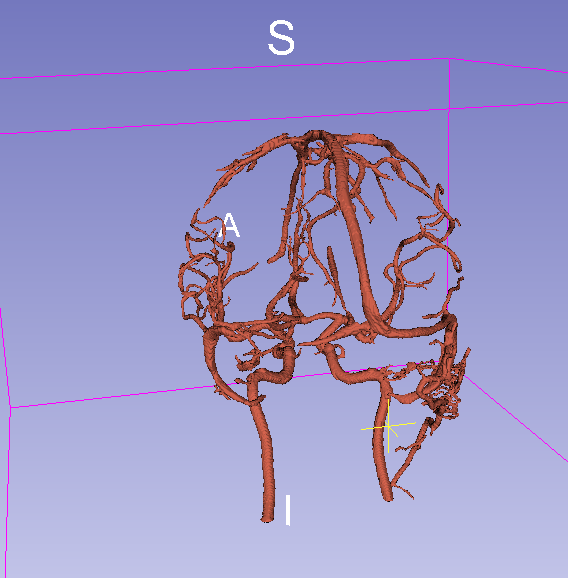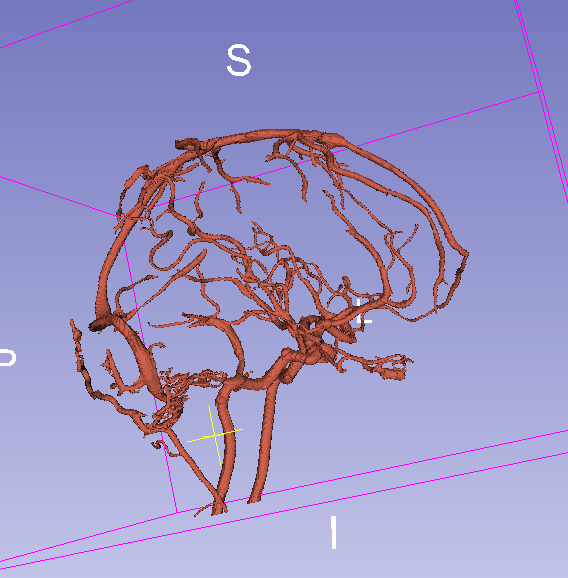
Final result with threshold = 0.07:

Final result with threshold = 0.15: